Research Overview
DNA repair proteins serve as the molecular caretakers of the genome. Mutations in these proteins can cause cancer and premature aging. We aim to unravel how these DNA repair proteins function at the molecular and cellular levels. To glimpse “under the hood” of these complex multi-protein reactions, we also develop high-throughput single-molecule and single-cell microscopy tools. For example, our “DNA curtain” approach combines aspects of surface chemistry, nanoengineering, microfluidics, and molecular biology to directly visualize how proteins function in real time. By observing these key reactions at the single-molecule level, we seek to understand the mechanisms of genome maintenance.
Ongoing research topics
How do DNA-binding molecular machines move on a crowded DNA molecule?
DNA binding proteins scan the genome for specific sites via facilitated diffusion along the DNA molecule. However, our genomes are crowded with other DNA-binding proteins (e.g., nucleosomes, transcription factors, etc). We are investigating how crowded DNA impacts facilitated diffusion and target recognition. For example, we are studying how the eukaryotic DNA mismatch repair (MMR) machinery navigates past nucleosomes to identify rare errors amidst a vast pool of error-free DNA. We are also exploring how the mismatch repair machinery is assembled at the lesion and how downstream mismatch repair is coupled to initial lesion recognition.
How do DNA repair proteins initiate double-strand breaks repair?
Physical breaks in the DNA duplex are the most toxic of all DNA lesions. Double-strand breaks occur when DNA replication fails or as a result of chemotherapeutic drugs and ionizing radiation. These DNA lesions are repaired by two major pathways: error-free homologous DNA recombination or error-prone non-homologous end joining (Fig. 1). We are currently exploring the first steps of DNA break repair by directly observing: (i) how human repair proteins locate broken DNA ends, (ii) how the broken DNA is directed into one of these two major repair pathways, (iii) and how repair proceeds on a highly condensed nucleosome-coated DNA.

Figure 1. Double-strand break (DSB) repair can proceed via error-prone non-homologous end joining (NHEJ) or error-free homologous recombination (HR).
How do defects in DNA repair lead to premature aging?
Aging--the progressive decline of function--is the leading risk factor for nearly every human disease. Defects in DNA repair pathways can accelerate aging at both the organismal and cellular levels. However, we still do not understand how cells avoid damage accumulation, renew cell vitality, or how dysregulation in these pathways leads to cell death. We use replicative aging in the fission yeast Schizosaccharomyces pombe (S. pombe) as a model system for understanding how DNA damage leads to premature aging in mitotically dividing cells (Fig. 2).
Specifically, we are investigating the links between genome maintenance, ribosomal DNA stability, and premature aging. A growing body of both cell biological and functional genomics studies conclude that S. pombe and human cells share similar biological pathways of DNA repair, cell cycle regulation, and mitochondrial maintenance. S. pombe is also amenable to both genetic and physical manipulation, and divides rapidly enough to tractably monitor all replication events for a given cell. For these reasons, we have developed a high-throughput microfluidic strategy for capturing and observing individual S. pombe cells over their entire lifespan (see below).
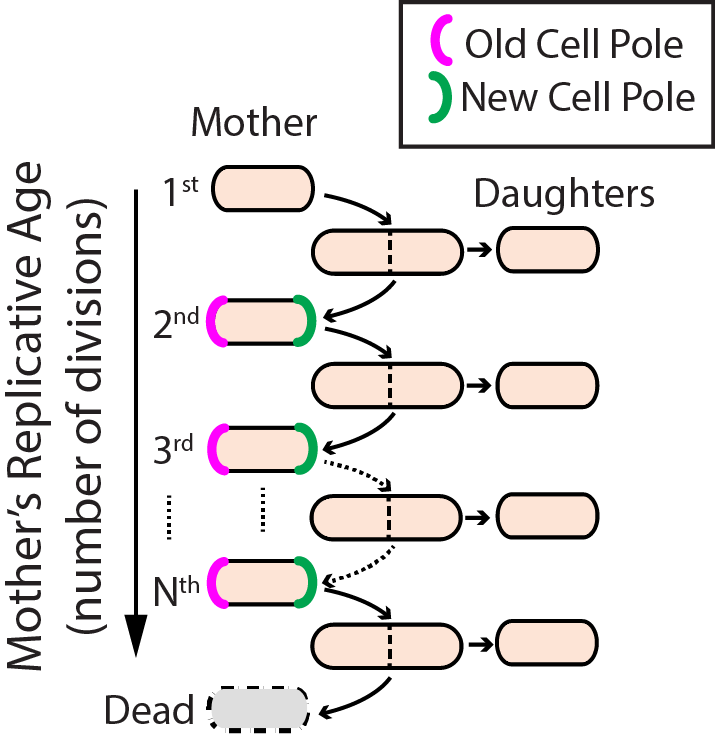
Figure 2. Replicative lifespan in fission yeast.
New high-throughput tools for single-molecule and single-cell research
Our lab uses Total Internal Reflection Fluorescence Microscopy (TIRFM) as the primary tool for single-molecule fluorescence imaging.
For TIRFM, laser excitation is limited to a shallow (~100 nanometer) penetration depth near the surface of a microfluidic flowcell. Molecules of interest are immobilized on a passivated flowcell surface, thus eliminating spurious background signals. Images are collected with a microscope objective and recorded using a back-illuminated charge-coupled device (CCD) with on-chip signal amplification. The CCD can be used with an image-splitter containing a dichroic mirror to separate the multicolor fluorescence signal for simultaneous multi-color imaging.

Figure 3. Total internal reflection fluorescence (TIRF) microscopy.
DNA Curtains
Collecting statistically relevant datasets is a major intrinsic challenge for experiments designed to observe individual reactions. We employ a high-throughput approach called “DNA curtains” that incorporates elements of nanofabrication and microfluidics to construct aligned arrays of surface-tethered DNA molecules. DNA curtains allow us to observe hundreds of biochemical reactions simultaneously at the single molecule level with excellent signal to noise and a wide choice of excitation and emission options. Furthermore, by nanofabricating various DNA curtain geometries, we can control critical experimental parameters such as DNA orientation, spacing, density and tension.
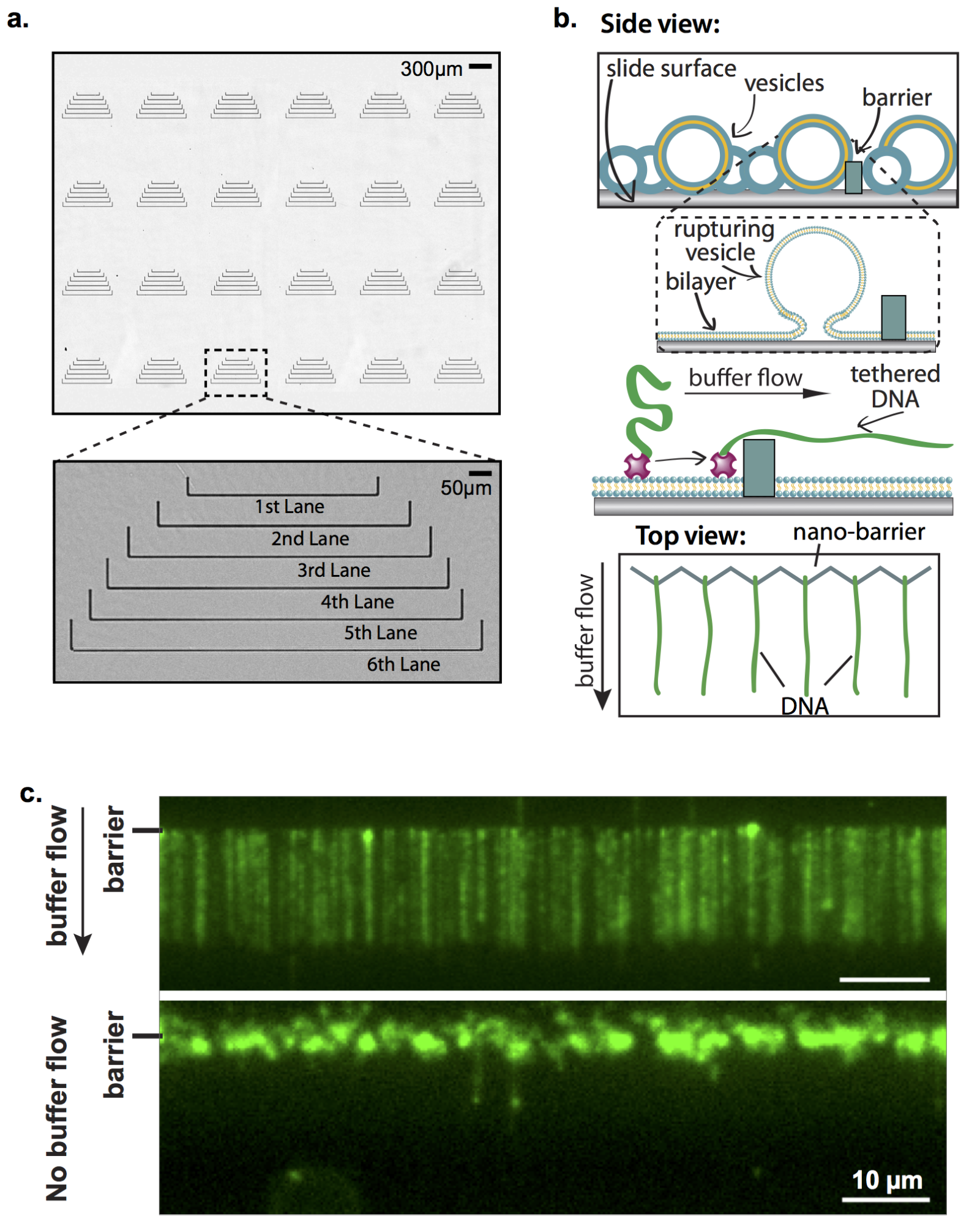
Figure 4. DNA Curtains: a high-throughput platform for single-molecule nucleic acid biochemistry.
Microfluidic single-cell capture technologies
Replicative lifespan studies require the microdissection of progeny cells, often for dozens of generations. These studies are laborious, low-throughput, and have largely remained unchanged since their introduction in 1959. Manual microdissection is especially challenging when the sibling cells are phenotypically similar, as occurs during fission yeast division. To address these challenges, we have developed the Fission Yeast Lifespan Microdissector (FYLM), a high-throughput microfluidic cell capture and microdissection device. With the FYLM, we can concurrently image the entire lifespan of thousands of individually addressable cells for over a week, while still maintaining sub-minute temporal resolution. Each cell is continuously supplied with fresh media, and imaged with subcellular resolution using brightfield and fluorescent microscopy.

Figure 5. Multi-day single cell capture and imaging in the fission yeast lifespan microdissector (FYLM).
We have also developed a semi-automated image analysis pipeline that permits quantitative analysis of each cell’s shape, doubling time, and fluorescent reporters. Like flow cytometry, this methodology yields single-cell level analysis in addition to population analysis, and cells can be extracted from the device for downstream manipulation. By combining our high-throughput FYLM technology with comprehensive lifespan analysis, this platform offers a unique approach to study long-lived mitotically active cells.





